G418 Disulfate, EvoPure® is a highly pure (≥99.0%) version of G418 Disulfate. It has been purified to remove the impurities commonly present in G418 Disulfate. This aminoglycoside antibiotic, originally isolated from Micromonospora rhodorangea, is routinely used for gene selection in cell culture.
We also offer:
- G418 Disulfate ReadyMadeTM Solution (G020-G021)
- G418 Disulfate (G001)
- G418 Disulfate (Low Endotoxin)(G048)
| Mechanism of Action | G418 Disulfate, and other aminoglycosides, including Kanamycin and Neomycin, prevent protein synthesis by blocking the elongation step in prokaryotic and eukaryotic ribosomes.
Mechanism of resistance: Resistance to G418 Disulfate is conferred by the neo gene (neomycin resistant gene) from either Tn5 or Tn601 (903) transposons. Cells successfully transfected with resistance plasmids containing the neo resistance gene can express aminoglycoside 3'-phosphotransferase (APT 3' I or APT 3' II) which covalently modifies G418 to 3-phosphoric G418. 3-phosphoric G418 has negligible potency and has low-affinity for prokaryotic or eukaryotic ribosomes. |
| Spectrum | G418 Disulfate is toxic to susceptible bacteria and fungi |
| Microbiology Applications | G418 Disulfate is used as a gene selection agent during transfection of eukaryotic cells. |
| Eukaryotic Cell Culture Applications | G418 Disulfate is routinely used as a selection agent in cell culture after transfection of eukaryotic cells. Resistant cells express the neo gene which produces aminoglycoside 3'-phosphotransferase (APT 3' I or APT 3' II), a protein that confers resistance to G418 Disulfate and other aminoglycoside antibiotics.
Optimal working concentrations:
The optimal working concentration of G418 Sulfate to select for resistant clones depends on the cell line, reagent quality, reagent lot, media, growth conditions, cell density, cell metabolic rate, cell cycle phase, and plasmid quality. A kill curve should be performed to determine the optimal concentration for each experimental system. Use the following guide to determine the concentration to use to generate a kill curve:
A working concentration of 200 mg/L is usually sufficient after resistant mammalian clones are selected and can be used for maintenance until stable resistant clones are selected. The Selectivity Factor is a quantifiable measure of how efficient an antibiotic is during the process of gene selection. Our technical team tested the selectivity factor of G418 for BHK-21 cells and HeLa cells. We found that G418 is an ideal selection antibiotic for transfected BHK-21 cells but not optimal for HeLa cells. The method uses a modified MTT assay, which can be used to numerically determine the antibiotic efficiency (Delrue I et al, 2018). For more information about the Selectivity Factor, click here. For more information on relevant cell lines, culture medium, and working concentrations, please visit the TOKU-E Cell-culture Database. |
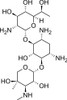
| Molecular Formula | C20H40N4O10 · xH2SO4 (lot specific) |
| Source | Biosynthetic: produced by Micromonospora rhodorangea. |
| Absorbance | 1mg/ml (water): 280nm <0.015 570nm <0.01 100mg/ml (water): 570nm <0.01 1.74g/25 ml (water): 280nm <0.7 |
| Documents | G418 Protocol.pdf|Plasmid_DNA_Transfection_Protocol.pdf|Selection_of_Stable_Transfected_Cell_Lines_Protocol.pdf |
| References |
Aragão FJL and Brasileiro ACM (2002) Positive, negative and marker-free strategies for transgenic plant selection. Braz. J. Plant Physiol. 14(1):1-10 Davis BD (1987) Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51(3):341-50 Lin-Cereghino J et al (2008) Direct selection of Pichia pastoris expression strains using new G418 resistance vectors. Yeast 25:293-299 Shin Y (2007) Selection of NptII transgenic sweet potato plants Using G418 and paromomycin. J. Plant Biol. 50(2):206-212 |
| Protocols |
G418 Disulfate Kill Curve ProtocolBackground: G418 Disulfate (syn: G418 Sulfate; Geneticin) is routinely used to select for successfully transfected mammalian cells that express the neo resistance gene in addition to the gene of interest. The neo gene encodes amino-glycoside 3’-phosphotransferase; an enzyme which confers resistance to G418 Disulfate and Neomycin. Before stable transfected cell lines can be selected, the optimal G418 Disulfate concentration needs to be determined by performing a kill curve titration. The optimal concentration of G418 Disulfate suitable for selection of resistant mammalian clones depends on the cell lines, media, growth conditions, and the quality of G418 Disulfate. It is necessary to perform a kill curve for every new cell type and new batch of G418 Disulfate. Preparation and storage of G418 disulfate solution:
Plasmid DNA Transfection Protocol
Once the appropriate antibiotic concentration to use for selection of the stable transfected cells has been determined by performing a kill curve, the next step is to generate a stable cell line by transfection of the parental cell line with a plasmid containing the gene of interest and an antibiotic resistance gene.
Plasmid DNA Transfection Protocol:
QC Seed 24-wells with insert and determine the transfection efficiency by immunostaining:
Selection of Stable Transfected Cell Lines ProtocolBackground: Once the cells have been successfully transfected, the next step is to seed and select the transfected cell line in a single 96-well plate to select pure colonies by limited dilution as outlined below: Protocol:
QC Seed 24-wells with insert for an immunostaining to determine percentage of cells expressing the gene of interest to be able to identify a 100% pure clone. You can also use Western blotting, flow cytometry or another technique depending on the cell line used. Seed 24-wells with insert and determine the expression level of the gene of interest by immunostaining:
|
| MIC | Enterobacter| 1 - >64| Escherichia coli (animal health isolate)| 1 - >64| Klebsiella| 1 - 32| |