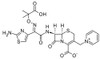
Ceftazidime Pentahydrate is a broad-spectrum, third-generation, β-lactam cephalosporin that interferes with bacterial cell wall synthesis. Patented in 1979 by Glaxo Group, Ceftazidime came into commercial use in 1984. Ceftazidime is bactericidal, exerting its effect by inhibition of enzymes responsible for cell-wall synthesis, primarily penicillin binding protein 3 (PBP3). Ceftazidime Pentahydrate is sparingly soluble in aqueous solution.
We also offer:
- Ceftazidime Hydrochloride (C020)
- Ceftazidime Solubilized (C019)
- Ceftazidime, Delta-3-Isomer, EvoPure (C121)
| Application | Ceftazidime Pentahydrate is used to study the expression of penicillin-binding proteins, since it disrupts the peptidoglycan layer in bacterial cell walls. |
| Mechanism of Action | Like β-lactams, cephalosporins interfere with PBP (penicillin binding protein) activity involved in the final phase of peptidoglycan synthesis. PBP’s are enzymes which catalyze a pentaglycine crosslink between alanine and lysine residues providing additional strength to the cell wall. Without a pentaglycine crosslink, the integrity of the cell wall is severely compromised and ultimately leads to cell lysis and death. Resistance to cephalosporins is commonly due to cells containing plasmid-encoded β-lactamases, however, Ceftazidime is stable in the presence of β-lactamases. |
| Spectrum | Ceftazidime Pentahydrate is broad-spectrum, targeting both Gram-negative and Gram-positive bacteria, but is most effective for Gram-negative strains including Pseudomonas aeruginosa and Enterobacteriaceae (including β-lactamase positive strains). It is also used against Streptococcus pneumoniae, and S. pyogenes. |
| Impurity Profile | Impurity A| delta-2-ceftazidime; ?(2RS,6R,7R)-7-[[(2Z)-2-(2-aminothiazol-4-yl)-2-[(1-carboxy-1-methylethoxy)imino]acetyl]amino]-8-oxo-3-[(1-pyridinio)methyl]-5-thia-1-azabicyclo[4.2.0]oct-3-ene-2-carboxylate||C22H24N6O7S2|548.60| Impurity B| (6R,7R)-7-[[(E)-2-(2-aminothiazol-4-yl)-2-[(1-carboxy-1-methylethoxy)imino]acetyl]amino]-8-oxo-3-[(1-pyridinio)methyl]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate||C22H22N6O7S2|546.58| Impurity C| (6R,7R)-2-carboxy-8-oxo-3-(pyridiniomethyl)-5-thia-1-azabicyclo[4.2.0]oct-2-ene-7-aminium dichloride||C13H13N3O3S|291.33| Impurity D| (6R,7R)-7-[[(Z)-2-[[2-(1,1-dimethylethoxy)-1,1-dimethyl-2-oxoethoxy]imino]-2-[2-[(triphenylmethyl)amino]thiazol-4-yl]acetyl]amino]-8-oxo-3-(pyridiniomethyl)-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate|||| Impurity E| (6R,7R)-7-[[(2Z)-2-(2-aminothiazol-4-yl)-2-[[2-(1,1-dimethylethoxy)-1,1-dimethyl-2-oxoethoxy]imino]acetyl]amino]-8-oxo-3-[(1-pyridinio)methyl]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate||C26H30N6O7S2|602.69| Impurity F| Pyridine|110-86-1|C5H5N|79.10| Impurity G| 2-[[[(1Z)-1-(2-aminothiazol-4-yl)-2-[(oxoethyl)amino]-2-oxoethylidene]amino]oxy]-2-methylpropanoic acid||C11H14N4O5S|314.32| Impurity H| (6R,7R)-7-[[(2Z)-2-(2-aminothiazol-4-yl)-2-[(2-methoxy-1,1-dimethyl-2-oxoethoxy)imino]acetyl]amino]-8-oxo-3-[(1-pyridinio)methyl]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate||C23H24N6O7S2|560.61| Ceftazidime| Delta-3-IsoMer||C22H22N6O7S25H2O|636.65| |
| Microbiology Applications | Ceftazidime Pentahydrate is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram negative microbial isolates. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
Media SupplementsCeftazidime can be used as a selective agent in several types of isolation media: PALCAM Agar - PALCAM Selective Supplement Chromogenic Listeria Agar - Chromogenic Listeria Selective Supplement Chromogenic Listeria Agar - Chromogenic Listeria Differential Supplement |
| Eukaryotic Cell Culture Applications | Human bronchial epithelial cells from chronically infected cystic fibrosis airways were cultured and exposed to Ceftazidime in order to analyze the cytotoxicity. Even at concentrations many times the antimicrobial level, the compound was nontoxic (Randell et al, 2001).
Ceftazidime present in aqueous solutions generate Cefazidime degradation products (CDPs) that inhibit in vitro HIV-1 replication in cell lines (MT-2) transformed with HTLV-I virions, CEM-SS, H9/HTLV-IIIB NIH 1983, Molt-4 clone 8, and U937 and in primary cells (lymphocytes and macrophages from peripheral blood mononuclear cells). The chemical nature of the active component responsible for the anti-HIV activity is currently unknown other than it is a high molecular weight polymer (MW 8000). The inhibitory action was manifested during the early phase of the HIV-1 lifecycle (Hobi et al, 2001). |
| Molecular Formula | C22H22N6O7S2 •5H2O |
| Impurities | Limit of Pyridine: ≤0.12% Ceftazidime Polymer: ≤0.3% |
| References |
Fischer J and Ganellin R (2006). Analogue-based drug discovery. John Wiley & Sons. p. 495 Hsu Y et al (2014) Biodegradable drug-eluting nanofiber-enveloped implants for sustained release of high bactericidal concentrations of vancomycin and Ceftazidime: In vitro and in vivo studies. Int. J. Nanomed. 9:4347-4355 Randell SH, Walstad DL, Schwab UE, Grubb BR and Yankaskas JR (2001) Isolation and culture of airway epithelial cells from chronically infected human lungs. In Vitro Cell. Dev. Biol.-Animal 37: 480 |
| MIC | Aeromonas spp.| 0.06 - 0.25|| Aeromonas spp. (ampicillin-resistant + cefazolin-resistant)| 0.12 - 4|| Agrobacterium radiobacter| 4 - 64|| Alcaligenes faecalis| 2 - 16|| Bacillus spp.| 0.5 - >256|| Bacteroides disiens| 4 - 16|| Bacteroides distasonis| 32 - >256|| Bacteroides fragilis| 4 - >256|| Bacteroides melaninogenicus | 0.125 - >64|| Bacteroides oralis | 1 - 32|| Bacteroides ovatus (ampicillin-resistant + cefazolin-resistant)| 32 - >64|| Bacteroides ruminicola subsp. brevis| 2 - >256|| Bacteroides septicum (ampicillin-resistant + cefazolin-resistant)| 32 - >64|| Bacteroides thetaiotaomicron| 4 - >256|| Bacteroides ureolyticus| ≤0.062 - 4|| Bordetella bronchiseptica| 1 - 32|| Bordetella pertussis | 0.125 - 0.5|| Brevundimonas vesicularis| 0.5 - >64|| Burkholderia cepacia| 1 - >512|| Burkholderia spp.| 0.25 - 8|| Campylobacter jejuni| 2 - 64|| Chryseobacterium indologenes| 4 - 32|| Chryseobacterium meningosepticum| ≥64|| Citrobacter amalonaticus| ≤2 - >16|| Citrobacter braakii| ≤2 - >128|| Citrobacter diversus| 0.03 - >128|| Citrobacter farmeri| ≤2 - >128|| Citrobacter freundii| 0.03 - 200|| Citrobacter koseri| ≤0.25 - >16|| Citrobacter spp.| 0.25 - >128|| Citrobacter werkmanii | ≥128|| Clostridium difficile| 32 - 256|| Clostridium perfringens| 0.5 - >16|| Comamonas acidovorans| 0.5 - 4|| Corynebacterium spp.| 0.25 - >256|| Edwardsiella hoshinae | ≤0.03|| Edwardsiella ictaluri | ≤0.03 - 0.5|| Edwardsiella tarda| ≤0.03 - 0.5|| Enterobacter aerogenes| 0.015 - >128|| Enterobacter agglomerans| ≤0.25 - 128|| Enterobacter amnigenus| 0.25 - 128|| Enterobacter asburiae| ≤1 - 128|| Enterobacter cancerogenus| ≤1 - >16|| Enterobacter cloacae| ≤0.03 - >128|| Enterobacter gergoviae| ≤2 - >16|| Enterobacter hormaechei| ≤1 - >16|| Enterobacter intermedius| 2 - 128|| Enterobacter sakazakii| ≤0.25 - 128|| Enterobacter spp.| ≤0.25 - 128|| Enterobacter taylorae| ≤1 - >16|| Enterobacteriaceae| 0.004 - 256|| Enterococci| 0.012 - 128|| Enterococcus faecalis| 4 - >128|| Enterococcus faecium| 16 - >128|| Enterococcus hirae (ATCC 10541)| >64|| Enterococcus spp.| >256|| Escherichia coli| 0.015 - 512|| Eubacterium spp.| 0.125 - 8|| Haemolytic streptococci| 0.015 - ≤8|| Haemophilus spp.| 0.015 - 0.5|| Hafnia alvei| 0.25 - >8|| Helicobacter pylori| 0.05 - 3.13|| Klebsiella ornithinolytica| ≤1 - >16|| Klebsiella oxytoca| 0.03 - >16|| Klebsiella ozaenae| ≤2 - >16|| Klebsiella pneumonia| ≤0.03 - >128|| Klebsiella rhinoscleromatis| 2|| Klebsiella spp.| 0.12 - 256|| Klebsiella terrigena| ≤2 - >16|| Lactobacillus acidophilus (CRL 1251)| 100|| Lactobacillus gasseri (CRL 1259)| >100|| Lactobacillus johnsonii (1294)| >100|| Lactobacillus paracasei (CRL 1289)| >100|| Lactobacillus salivarius (CRL 1328)| 100|| Leptotrichia buccalis| 0.5|| Listeria monocytogenes (cefazolin-resistant)| 32 - >64|| Micrococcus spp.| 16 - 32|| Moraxella catarrhalis| 0.015 - >8|| Morganella morganii| 0.03 - 128|| Morganella spp.| ≤0.25 - 128|| Neisseria meningitidis| <0.13 - 0.25|| Neisseria spp.| 0.004 - 0.5|| Ochrobactrum anthropi| ≥64 - ≥128|| Oligella ureolytica| 0.5 - >64|| Pantoea agglomerans| 0.12 - >16|| Pasteurella multocida| 0.03 - 4|| Pemphigus mirabilis| <0.5 - 1|| Pemphigus vulgaris| 0.25 - 1|| Peptococcus asaccharolyticus| 0.5 - 16|| Peptostreptococcus anaerobius| 8 - 16|| Plesiomonas shigelloides| 0.03 - 4|| Pneumococci| 0.03 - 32|| Prevotella bivia| 16 - 32|| Prevotella melaninogenica | 8|| Proteae spp.| ≤2 - >16|| Proteus mirabilis| ≤0.007 - 99|| Proteus penneri| 0.12 - ≤2|| Proteus rettgeri| 0.03 - 128|| Proteus spp.| ≤0.06 - 8|| Proteus vulgaris| ≤0.03 - 128|| Providencia alcalifaciens (indole-positive)| 0.03 - 128|| Providencia rettgeri| 0.03 - >32|| Providencia spp.| ≤0.06 - 128|| Providencia stuartii| 0.03 - 128|| Pseudomonas aeruginosa| ≤0.03 - 1024|| Pseudomonas cepacia| 2 - >64|| Pseudomonas oryzihabitans| 0.13 - >64|| Pseudomonas spp.| 0.25 - 128|| Pseudomonas stutzeri| ≤0.06 - >64|| Ralstonia pickettii | 2 - 16|| Ralstonia pickettii (PIC-1)| 0.5|| Salmonella| ≤2 - >16|| Salmonella agona| ≤2 - >16|| Salmonella arizonae| ≤2 - >16|| Salmonella bareilly| ≤2 - >16|| Salmonella enterica| ≤2 - >16|| Salmonella enteritidis| 0.125 - >16|| Salmonella hadar| ≤2 - >16|| Salmonella heidelberg| ≤2 - >16|| Salmonella infantis| ≤2 - >16|| Salmonella litchfield| ≤2 - >16|| Salmonella Montevideo| ≤2 - >16|| Salmonella muenchen| ≤2 - >16|| Salmonella Newport| ≤2 - >16|| Salmonella panama| ≤2 - >16|| Salmonella Paratyphi| ≤2 - >16|| Salmonella schwarzengrund| ≤2 - >16|| Salmonella spp.| 0.12 - 16|| Salmonella stanley| ≤2 - >16|| Salmonella stpaul| ≤2 - >16|| Salmonella thompson| ≤2 - >16|| Salmonella typhi| 0.06 - >16|| Salmonella virchow| ≤2 - >16|| Serratia fonticola| ≤1 - 64|| Serratia liquefaciens| ≤1 - >16|| Serratia marcescens| ≤0.03 - >128|| Serratia odorifera| 0.25 - >16|| Serratia plymuthica| ≤1 - >16|| Serratia rubidaea| ≤1 - 64|| Serratia spp.| ≤0.25 - 128|| Shigella boydii| ≤2 - 4|| Shigella dysenteriae| 0.125 - 4|| Shigella flexneri| 0.06 - 64|| Shigella sonnei| 0.06 - 4|| Shigella spp.| 0.06 - 4|| Sphingomonas spp.| 2 - >64|| Staphylococci| 0.06 - >256|| Staphylococcus aureus| ≤0.12 - 512|| Staphylococcus auricularis (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Staphylococcus capitis (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Staphylococcus caprae (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Staphylococcus cohnii (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Staphylococcus epidermidis | 0.25 - 256|| Staphylococcus haemolyticus| 0.25 - >128|| Staphylococcus hominis| 0.25 - >128|| Staphylococcus intermedius (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Staphylococcus lugdunensis (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Staphylococcus mitis| 0.06 - 64|| Staphylococcus sanguis| 0.03 - 4|| Staphylococcus saprophyticus| 0.25 - >64|| Staphylococcus sciuri | 0.25 - >16|| Staphylococcus simulans| 0.25 - >128|| Staphylococcus spp.| 0.25 - >16|| Staphylococcus warneri (coagulase-negative + oxacillin-susceptible)| 0.25 - >16|| Stenotrophomonas maltophilia| 0.5 - >256|| Streptococci| 0.12 - 16|| Streptococcus agalactiae| 0.12 - 0.5|| Streptococcus pneumonia| 0.015 - 32|| Streptococcus pyogenes| 0.015 - 0.5|| Vibrio cholerae| 5 - 60|| Vibrio parahaemolyticus (KX-V138)| 10|| Weeksella virosa | 0.25 - 8|| Xanthomonas maltophilia| 2 - >64|| Yersinia enterocolitica| 0.03 - 8|| |