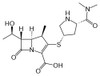
Meropenem is a β-lactam antibiotic in the carbapenem class, and targets the bacterial cell wall. It has found utility against extended spectrum β-lactamase (ESBL) producing Enterobacteriaceae that are resistant to many first line β-lactam antibiotics and certain cephalosporins. Meropenem is sparingly soluble in aqueous solution.
We also offer: