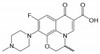
Ofloxacin is a synthetic, broad-spectrum, second-generation fluoroquinolone antibiotic that is an analog of nalidixic acid. It was developed by Daiichi Seiyaku to be resistant to metabolic oxidation in the liver and inhibits the supercoiling activity of bacterial DNA gyrase, halting DNA replication. It is used to combat organisms causing urinary tract and respiratory infections.
| Mechanism of Action | Ofloxacin is a Cytochrome P-450 inhibitor and DNA Topoisomerase II inhibitor. Fluoroquinolone antibiotics target bacterial DNA gyrase, an enzyme which reduces DNA strain during replication. Because DNA gyrase is required during DNA replication, subsequent DNA synthesis and ultimately cell division is inhibited. |
| Spectrum | Ofloxacin is a broad-spectrum antibiotic targeting a wide range of Gram-positive and Gram-negative organisms. |
| Impurity Profile | Impurity A| (RS)-9,10-Difluoro-3-methyl-7-oxo-2,3-dihydro-7H-pyrido[1,2,3-de]-1,4-benzoxazine-6-carboxylic Acid (FPA) ; Ofloxacin Difluoro Carboxylic Acid|82419-35-0|C13H9F2NO4|281.21| Impurity B| (RS)-9-Fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-2,3-dihydro-7H-pyrido-[1,2,3-de]-1,4-benzoxazin-7-one|||| Impurity E| (RS)-9-Fluoro-3-methyl-7-oxo-10-(piperazin-1-yl)-2,3-dihydro-7H-pyrido[1,2,3-de]-1,4-benzoxazine-6-carboxylic Acid; N-Desmethyl Ofloxacin HCl|82419-52-1|C17H18FN3O4|347.34| Impurity F| 4-[(RS)-6-Carboxy-9-fluoro-3-methyl-7-oxo-2,3-dihydro-7H-pyrido[1,2,3-de]-1,4-benzoxazine-10-yl]-1-methylpiperazine 1-Oxide Hydrochloride (Ofloxacin N-Oxide Acetate Salt)|104721-52-0|C20H24FN3O7|437.42| | Ofloxacin R-Isomer; (+)-(R)-9-Fluoro-2,3-dihydro-3-methyl-10-(4-methyl-1-piperazinyl)-7-oxo-7H-pyrido[1,2,3-de]-1,4-benzoxazine-6-carboxylic acid ;(R)-(+)-Ofloxacin ;D-Ofloxacin ; Levofloxacin R-Isome|100986-86-5|C18H20FN3O4|361.37| |
| Microbiology Applications | Ofloxacin is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram-positive and Gram-negative microbial isolates. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
For a representative list of Ofloxacin MIC values, click here. |
| Cancer Applications | Oflaxacin had an inhibitory effect on the proliferation of transitional cell carcinoma cell lines at high concentrations (>200 μg/ml). The effect may be related to impairment of telomerase activity by some unknown mechanism (Yamakuchi M et al, 1997).
Ofloxacin inhibited proliferation and DNA synthesis of these 3 human TCC lines (CCSUP, T24, and J82 ) in vitro. Inhibition occurred in a concentration- and time-dependent manner. Inhibition of proliferation and DNA synthesis were assessed via MTT and tritiated thymidine assays (Seay, 1996). |
| Molecular Formula | C18H20FN3O4 |
| Solubility | Glacial Acetic Acid: Soluble Methanol: Slightly soluble Water: Slightly soluble |
| References |
Mouton Y and Leroy O (1991) Ofloxacin. Int. J. Antimicrob. Agents 1(2-3):57-74 Seay TM, Peretsman SJ and Dixon PS (1996) Inhibition of human transitional cell carcinoma in vitro proliferation by fluoroquinolone antibiotics. J. Urol. 2:757-762 PMID 8558720 Smith JT (1991) Ofloxacin, a bactericidal antibacterial. Chemother. 37 (supp 1):2-13 PMID 1646700 Yamakuchi M et al (1997) New quinolones, Ofloxacin and levofloxacin, inhibit telomerase activity in transitional cell carcinoma cell lines. Cancer Lett. 119(2):213-219 PMID 9570374 |
| MIC | Bacillus cereus| ≥5|| Bacillus macerans| ≥15.62|| Bacillus subtilis| 0.05 - 125|| Bacteroides bivius| ≥0.78|| Bacteroides distasonis| 2 - 16|| Bacteroides fragilis| 1 - 16|| Bacteroides intermedius| ≥0.78|| Bacteroides oralis| ≥3.13|| Bacteroides oris| ≥0.78|| Bacteroides ovatus| 2 - 16|| Bacteroides thetaiotaomicron| 2 - 16|| Bacteroides uniformis| ≥6.25|| Bacteroides ureolyticus| 0.1 - 8|| Bacteroides vulgatus| 2 - 16|| Bifidobacterium adolescentis| ≤0.025|| Bordetella bronchiseptica| 0.5 - 2|| Bordetella pertussis | 0.063 - 0.125|| Borrelia burgdorferi S.L.| 1 - 16|| Brucella melitensis| <0.125 - >16|| Burkholderia cepacia| 0.125 - 32|| Burkholderia mallei| 64 - 128|| Capnocytophaga ochracea | ≤0.025|| Cedecea davisae| 62.5|| Chlamydia pneumonia| 0.5 - 2|| Chlamydia psittaci| 0.5|| Chlamydophila pneumonia| 1|| Citrobacter freundii| 0.031 - 128|| Citrobacter spp.| 0.25|| Clostridium cadaveris| 0.25 - >32|| Clostridium difficile| 8 - 12.5|| Clostridium innocuum| 0.25 - >32|| Clostridium malenominatum| 0.25 - >32|| Clostridium oroticum| 0.25 - >32|| Clostridium perfringens| 0.25 - >32|| Clostridium septicum| 0.39|| Clostridium sordellii| 0.25 - >32|| Clostridium sporogenes| 0.25 - >32|| Clostridium spp.| 8|| Corynebacterium diphtheriae| ≥0.2|| Corynebacterium jeikeium| 0.5 - 32|| Enterobacter aerogenes| 0.1 - 0.2|| Enterobacter cloacae| 0.031 - 16|| Enterobacter spp.| 0.25|| Enterobacteriaceae| 0.06 - 128|| Enterococci| 1 - 128|| Enterococcus avium| 2 - 4|| Enterococcus casseliflavus| 2 - 8|| Enterococcus faecalis| 0.78 - >128|| Enterococcus faecium| 0.39 - >128|| Enterococcus gallinarum| 2 - 4|| Enterococcus hirae| 0.39 - 6.25|| Enterococcus raffinosus| 2 - 4|| Erysipelothrix spp.| ≥0.25|| Escherichia coli| 0.0125 - 62.5|| Eubacterium| 0.12 - 8|| Eubacterium lentum| ≥0.78|| Eubacterium limosum| 0.12 - 8|| Eubacterium nodatum| 0.12 - 8|| Finegoldia magna| 0.25 - 32|| Fusobacterium mortiferum (GAI 5576)| ≥3.13|| Fusobacterium nucleatum| 1 - 2|| Fusobacterium varium (ATCC 8501)| ≥6.25|| Haemophilus influenzae| <0.03 - 0.25|| Haemophilus spp.| 0.015 - 2|| Hafnia alvei (NCTC 9540)| ≥0.1|| Helicobacter pylori| 0.05 - 25|| Klebsiella pneumonia| 0.063 - 125|| Klebsiella spp.| 0.5|| Lactobacillus acidophilus| 8 - 64|| Lactobacillus brevis| 2 - 16|| Lactobacillus casei| 0.5 - 64|| Lactobacillus curvatus| 8 - 16|| Lactobacillus delbrueckii| 4 - 32|| Lactobacillus fermentum| 8 - 16|| Lactobacillus jensenii| 0.12 - 8|| Lactobacillus plantarum| 8 - 32|| Lactobacillus rhamnosus| 0.5 - 64|| Lactobacillus sakei| 8 - 16|| Lactobacillus sp.| 0.12 - 128|| Legionella pneumophila| 0.03|| Leuconostoc spp.| 2 - 4|| Listeria monocytogenes| 1 - 2|| Micrococcus luteus (ATCC 9341)| ≥1.56|| Micromonas micros| 0.25 - 32|| Moraxella catarrhalis| ≤0.03 - 0.25|| Moraxella spp.| 0.19|| Morganella morganii| 0.063 - 128|| Mycobacterium africanum| 0.5|| Mycobacterium bovis| 1|| Mycobacterium tuberculosis| ≤0.125 - 16|| Mycoplasma fermentans| 0.06 - 0.25|| Mycoplasma genitalium| 0.5 - 2|| Mycoplasma hominis| 0.25 - 1|| Mycoplasma penetrans| 0.12 - 0.5|| Mycoplasma pneumonia| 0.5 - 2|| Neisseria cinerea| 0.015 - 0.25|| Neisseria lactamica| 0.015 - 0.03|| Neisseria meningitidis| 0.007 - 0.03|| Neisseria mucosa| 0.015 - 0.06|| Neisseria perflava| 0.015 - 0.12|| Neisseria polysaccharea| 0.007 - 0.015|| Neisseria sicca| 0.015 - 0.12|| Neisseria spp.| 0.001 - 0.06|| Pediococcus spp.| 8 - 16|| Peptostreptococcus| 0.25 - 32|| Peptostreptococcus anaerobius| 0.25 - 32|| Peptostreptococcus asaccharolyticus| 0.25 - 32|| Peptostreptococcus magnus| 0.39|| Peptostreptococcus prevotii| 0.25 - 32|| Peptostreptococcus spp.| 8|| Pneumococci| 1 - 128|| Prevotella bivia| 1 - 32|| Prevotella buccae| 1 - 32|| Prevotella denticola| 1 - 32|| Prevotella disiens| 1 - 32|| Prevotella intermedia| 1 - 32|| Prevotella loescheii| 1 - 32|| Prevotella melaninogenica| 1 - 32|| Prevotella oralis| 1 - 32|| Prevotella oris| 1 - 32|| Prevotella spp.| 1 - 32|| Propionibacterium| 0.12 - 8|| Proteus mirabilis| 0.063 - 0.5|| Proteus spp.| ≥0.5|| Proteus vulgaris| 0.031 - 125|| Providencia inconstans| 0.39|| Providencia rettgeri| 0.063 - >128|| Pseudomonas aeruginosa| 0.5 - >128|| Pseudomonas syringae| ≥125|| Salmonella enteritidis| ≥0.05|| Salmonella paratyphi| 0.025 - 0.05|| Salmonella spp.| 0.12|| Salmonella typhi| 0.012 - 0.025|| Serratia marcescens| 0.125 - 32|| Shigella| 0.003 - 1|| Shigella boydii| ≥0.1|| Shigella dysenteriae| ≥0.05|| Shigella flexneri | 0.05 - 0.1|| Shigella sonnei| 0.05 - 0.06|| Shigella spp.| 0.06|| Staphylococci| 0.12 - 128|| Staphylococcus asaccharolyticus| ≥0.78|| Staphylococcus aureus| 0.12 - 128|| Staphylococcus auricularis| ≤0.12 - >30|| Staphylococcus capitis| ≤0.12 - >30|| Staphylococcus epidermidis| 0.063 - >128|| Staphylococcus faecalis| ≥5|| Staphylococcus haemolyticus| ≤0.12 - >64|| Staphylococcus hominis| ≤0.12 - >30|| Staphylococcus intermedius| ≤0.12 - >30|| Staphylococcus lugdunensis| ≤0.12 - >30|| Staphylococcus pyogenes| 0.5 - 2|| Staphylococcus saprophyticus| ≤0.12 - >30|| Staphylococcus simulans| ≤0.12 - >30|| Staphylococcus warneri| ≤0.12 - >30|| Stenotrophomonas maltophilia| ≤0.25 - 32|| Streptococci| 0.5 - 64|| Streptococcus agalactiae| 1 - 2|| Streptococcus faecalis| ≥4|| Streptococcus intermedius | 1.56|| Streptococcus pneumonia| 0.5 - 256|| Streptococcus pyogenes| 0.78 - 62.5|| Streptococcus spp.| 1.56 - 4|| Ureaplasma spp.| 0.5 - 2|| Ureaplasma urealyticum| 0.5 - 2|| Veillonella parvula| ≥0.39|| Veillonella spp.| 0.25 - 16|| Yersinia enterocolitica| ≥62.5|| |