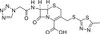
Cefazolin Sodium, USP is a first generation cephalosporin. It is a semi-synthetic, broad-spectrum β-lactam with bactericidal activity. It can be used to study penicillin binding proteins, especially PBP2 and PBP4 on the synthesis of bacterial cell walls.
Cefazolin Sodium is sparingly soluble in aqueous solution.
| Mechanism of Action | Like β-lactams, cephalosporins interfere with PBP (penicillin binding protein) activity involved in the final phase of peptidoglycan synthesis. PBP’s are enzymes which catalyze a pentaglycine crosslink between alanine and lysine residues providing additional strength to the cell wall. Without a pentaglycine crosslink, the integrity of the cell wall is severely compromised and ultimately leads to cell lysis and death. Resistance to cephalosporins is commonly due to cells containing plasmid encoded β-lactamases. |
| Spectrum | Cefazolin Sodium is effective against Gram-positive and Gram-negative bacteria. |
| Impurity Profile | Impurity A| (6R,7R)-7-amino-3-[[(5-methyl-1,3,4-thiadiazol-2-yl)sulphanyl]methyl]-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid|30246-33-4|C11H12N4O3S3|344.44| Impurity B| (6R,7R)-7-[(2,2-dimethylpropanoyl)amino]-3-[[(5-methyl-1,3,4-thiadiazol-2-yl)sulphanyl]methyl]-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid||C16H20N4O4S3|428.55| Impurity C| (6R,7R)-3-methyl-8-oxo-7-[(1H-tetrazol-1-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid|56842-77-4|C11H12N6O4S|324.32| Impurity D| (6R,7R)-3-[(acetyloxy)methyl]-8-oxo-7-[(1H-tetrazol-1-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid|32510-61-5|C13H14N6O6S|382.36| Impurity E| MMTD|29490-19-5|C3H4N2S2|132.2| Impurity G| (5aR,6R)-6-[(1H-tetazol-1-ylacetyl)amino-5a,6-dihydro 3H,7H-azeto[2,1-b]furo[3,4-d][1,3]thiazine-1,7(4H)-dione||C11H10N6O4S|322.30| Impurity H| 7-ACA|957-68-6|C10H12N2O5S|272.28| Impurity I| cefazoloic acid; 2-[carboxy[(1H-tetrazol-1-ylacetyl)amino]methyl]-5-[[(5-methyl-1,3,4-thiadiazol-2-yl)sulfanyl]methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid||C14H16N8O5S3|472.53| Impurity J| hydrolysed cefazoloic acid; 2-[carboxy[(1H-tetrazol-1-ylacetyl)amino]methyl]-5-(hydroxymethyl)-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid||C11H14N6O6S|358.33| Impurity K| Cefazolinamide; (6R,7R)-3-[[(5-methyl-1,3,4-thiadiazol-2-yl)sulfanyl]methyl]-8-oxo-7-[(1H-tetrazol-1-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxamide||C14H15N9O3S3|453.53| Impurity L| (6R,7S)-3-[[(5-methyl-1,3,4-thiadiazol-2-yl)sulphanyl]methyl]-8-oxo-7-[(1H-tetrazol-1-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid 1H-Tetrazole-1-acetic acid|21732-17-2|C3H4N4O2|128.09| |
| Microbiology Applications | Cefazolin Sodium is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram-positive and Gram-negative microbial isolates. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
Media SupplementCefazolin Sodium is routinely used as a selection agent in isolation media: Columbia Blood Agar - Campylobacter selective supplement (Butzler) |
| Eukaryotic Cell Culture Applications | The reducing power of the cell involves Glucose-6-phosphate dehydrogenase (G6PD). The reprograming of nicotinamide adenine dinucleotide (NAD) homeostasis is reported to be an important step in cancer progression. Rat erythrocyte G6PD enzyme was studied in vitro with Cefazolin and it was found that the enzyme increased upon exposure to Cefazolin (Temel et al, 2018). The toxicity of cephalosporin to rabbit kidney cells in vitro was compared to their known nephrotoxic potential in vivo. Researchers found that when they added a microsomal S9 fraction from rabit kidney was added to the culture, Cephalothin was desacetylated and its toxicity to kidney cells was reduced. Desacetyl‐cephalothin was relatively nontoxic to rabbit renal tissue in vitro (Hottendorf et al, 1987). |
| Molecular Formula | C14H13N8NaO4S3 |
| References | Georgopapadakou, NH (1992) Mechanisms of action of Cephalosporin 3'-quinolone esters, carbamates, and tertiary amines in Escherichia coli. Antimicrob. Agents Chemother. 37(3):559-565 Hottendorf GH, Laska DA, Williams PD and Ford SM (1987) Role of desacetylation in the detoxification of Cephalothin in renal cells in culture. J Toxicol. Environ. Health 22(1): 101-111 PMID 3612832 Pedroso TM and Salgado HRN (2014) Development and validation of a microbiological assay by turbidimetry to determine the potency of Cefazolin Sodium in the lyophilized powder form. Anal. Methods 6:1391-1396 Reller LB, Karney WW, Beaty HN, Holmes KK, Turck M (1973) Evaluation of Cefazolin, a new cephalosporin antibiotic. Antimicrob Agents Chemother. 3(4):488-497 PMID 4790605 Ries K, Matthew E. Levison ME and Kaye D (1973) Clinical and In vitro evaluation of Cefazolin, a new Cephalosporin antibiotic. Antimicrob. Agents Chemother. 3 (2) 168-174 PMID 4790605 Temel Y, Ayna A, Shafeeq IH and Ciftci M (2018) In vitro effects of some antibiotics on glucose-6-phosphate dehydrogenase from rat (Rattus norvegicus) erythrocyte. Drug and Chem. Toxicol. 27:1-5 PMID 29947262 |
| MIC | Bacteroides fragilis| 64|| Bacteroides fragilis (ATCC 25285)| 64|| Bacteroides fragilis (UC-2)| 16|| Bacteroides thetaiotaomicron| 16|| Citrobacter diversus| 3.5|| Citrobacter freundii| 10 - 25.4|| Citrobacter freundii (Basal amp(C))| 8|| Citrobacter freundii (Hyperproducing amp(C))| 2048|| Citrobacter freundii (Inducible amp(C))| 16|| Citrobacter spp.| 0.5 - >128|| Clostridium perfringens| 0.5|| Clostridium perfringens (ATCC 13124)| 1|| Clostridium ramosum| 4|| Enterobacter| 50 - 400|| Enterobacter aerogenes| 8 - >128|| Enterobacter agglomerans| 1 - 64|| Enterobacter cloacae| 1 - >512|| Enterobacter cloacae (A-5053)| >128|| Enterobacter cloacae (A-5140)| >128|| Enterobacter cloacae (Basal amp(C))| 16|| Enterobacter cloacae (Hyperproducing amp(C))| 2048|| Enterobacter cloacae (Inducible amp(C))| 2048|| Enterobacter spp.| 1 - >128|| Escherichia coli| 0.5 - >412.8|| Escherichia coli (ATCC 25922)| 8|| Escherichia coli (ATCC 35218)| 64|| Escherichia coli (Basal amp(C))| 8|| Escherichia coli (cephalothin-resistant)| 0.8 - 400|| Escherichia coli (Hyperproducing amp(C))| 512|| Escherichia coli (Juhl)| 1.6|| Eubacterium lentum| 64|| Fusobacterium (penicillin-susceptible)| ≤0.015 - 8|| Fusobacterium mortiferum| 128|| Fusobacterium necrophorum| 0.06|| Haemophilus influenzae| 0.03 - 16|| Klebsiella| 0.8 - 50|| Klebsiella baumannii| >512|| Klebsiella oxytoca| 4.2|| Klebsiella pneumonia| 1 - >512|| Klebsiella pneumonia (ESBL)| >256|| Klebsiella pneumonia (SHV-1)| 8|| Klebsiella pneumonia (β-lactamase negative)| 8|| Klebsiella spp.| 1 - >128|| Leuconostoc spp.| 16000|| Listeria monocytogenes| 0.67|| Morganella morganii (Hyperproducing amp(C))| >2048|| Morganella morganii (Inducible amp(C))| >2048|| Neisseria meningitidis| 0.25 - 0.5|| Nocardia asteroides| 12.5 - ≥400|| Pandoraea apista| 2 - >32|| Pandoraea genomo| >32 - ?|| Pandoraea pnomenusa| >32 - ?|| Pasteurella multocida| <2 - ?|| Peptococcus asaccharolyticus (ATCC 29743)| 0.5 - ?|| Peptococcus magnus | 0.25 - ?|| Peptostreptococcus (penicillin-resistant)| ≤0.015 - 8|| Peptostreptococcus (penicillin-susceptible)| ≤0.015 - 2|| Porphyromonas (penicillin-susceptible)| ≤0.015 - 2|| Proteus mirabilis| 1 - 97|| Proteus morganii| 95.1 - ?|| Proteus rettgeri| 38.6 - ?|| Proteus spp. (indole-positive)| 0.25 - >129|| Proteus spp. (indole-positive)| 0.5 - >128|| Proteus vulgaris| 44.2 - 50|| Proteus vulgaris (Hyperproducing amp(C))| 512|| Proteus vulgaris (Inducible amp(C))| 512|| Providencia| 50 - 400|| Providencia spp.| 8 - 128|| Providencia spp. (indole-positive)| 0.5 - >128|| Pseudomonas aeruginosa| >128 - >512|| Pseudomonas aeruginosa (A-5005)| >128|| Pseudomonas aeruginosa (A-5007)| >128|| Pseudomonas aeruginosa (ATCC 27853)| >512|| Pseudomonas spp.| >128 - >256|| Salmonella| 0.8 - 100|| Serratia marcescens| 16 - >256|| Serratia marcescens (Hyperproducing amp(C))| >2048|| Serratia marcescens (Inducible amp(C))| >2048|| Shigella| 0.8 - 200|| Shigella flexneri| 0.5 - 32|| Shigella sonnei| 1 - 4|| Shigella spp.| 2|| Staphylococcus aureus| <0.2 - >344|| Staphylococcus epidermidis| ≤0.06 - >128|| Staphylococcus epidermidis (methicillin-susceptible)| 0.25 - 4|| Staphylococcus haemolyticus (methicillin-resistant)| 0.25 - 4|| Staphylococcus intermedius (ATCC 29663)| 0.063|| Staphylococcus pseudintermedius| ≤0.06 - 0.25|| Staphylococcus pyogenes| 0.12|| Staphylococcus saprophyticus (methicillin-susceptible)| 0.25 - 4|| Streptococci (group D)| 0.25 - 64|| Streptococci (Viridans group + penicillin-resistant)| 2 - 4|| Streptococci (Viridans group + penicillin-susceptible)| ≤0.015 - 4|| Streptococcus (group C)| 0.06 - 8|| Streptococcus agalactiae (group B)| 0.125 - 0.25|| Streptococcus bovis| 0.06 - 8|| Streptococcus pneumonia| 0.125 - 64|| Streptococcus pneumonia (SP-4 + penicillin-resistant)| 8|| Streptococcus pneumoniae| 0.03 - 0.12|| Streptococcus pyogenes (group A)| 0.125 - 0.25|| Streptococcus spp. (Viridans group)| 0.06 - 8|| Treponema hyodysenteriae | 6.25 - 50|| |