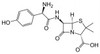
Amoxicillin Sodium is an extended spectrum β-lactam antibiotic and is similar in structure to Ampicillin. Resistance to Amoxicillin can be attributed to β-lactamase enzymes secreted by a resistant cell. Amoxicillin Sodium is soluble in aqueous solution.
We also offer:
| Mechanism of Action | Like all β-lactams, Amoxicillin targets PBP’s (penicillin binding proteins) involved in the final phase of peptidoglycan synthesis. PBP’s are enzymes which catalyze a pentaglycine crosslink between alanine and lysine residues. Without a pentaglycine crosslink, the integrity of the cell wall is severely compromised ultimately leading to the death of the cell. |
| Spectrum | Amoxicillin targets a wide range of β-lactamase negative Gram-positive and Gram-negative bacteria including E. coli and a number of Streptococcus and Staphylococcus species. Because peptidoglycan is synthesized in Gram-positive and Gram-negative bacteria, Amoxicillin can be used against a wide variety of species. |
| Microbiology Applications | Amoxicillin is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram-positive and Gram-negative microbial isolates. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
|
| Eukaryotic Cell Culture Applications | Dendritic cells were incubated with supernatant from Amoxicillin-treated hepatocytes in vitro to characterize signaling pathways between hepatocytes and immune cells to study the compound's influence on the immune response (Ogese, 2017). |
| Molecular Formula | C16H18N3NaO5S |
| Impurities | Related Substances Amoxicillin dimer: ≤3.0% Any impurity: ≤3.0% Sum of impurities: ≤9.0% 2-Ethyl hexanoic acid: ≤1.0% |
| Residual Solvents | Ethanol: ≤0.5% Methyl acetate: ≤0.5% |
| References | Ogese MO (2017) Characterization of drug-specific signaling between primary human hepatocytes and immune cells. Toxicol Sci. 158(1):76-89 PMID 28444390 Pitout JD, Sanders CC, Sanders WE (1997) Antimicrobial resistance with focus on beta-lactam resistance in gram-negative bacilli. Am J Med 103(1):51-59. PMID 9236486 Worlitzch D et al (2001) Effects of Amoxicillin, gentamicin, and moxifloxacin on the hemolytic activity of Staphylococcus aureus in vitro and in vivo. Antimicrob Agents Chemother. 45(1):196-202 PMID 11120965</em<> |
| MIC | Aggregatibacter actinomycetemcomitans| 1 - 16|| Bacillus cereus| 4 - 31.25|| Bacillus pumilu| 1|| Bacillus subtilis| 0.05 - 412|| Bacteroides distasonis| 2 - >128|| Bacteroides fragilis| ≤0.25 - ≥512|| Bacteroides ovatus| 16 - >128|| Bacteroides thetaiotaomicron| 16 - >128|| Bacteroides uniformis| 16 - >128|| Bacteroides vulgatus| 8 - >128|| Bifidobacterium spp.| ≤0.125 - 0.5|| Borrelia burgdorferi S.L.| 0.03 - 2|| Burkholderia cepacia (8Q)| >18|| Burkholderia mallei (Pakistan)| 128|| Campylobacter gracilis (ATCC 33236)| 1|| Campylobacter helveticus (ATCC 51209)| 4|| Campylobacter jejuni| 0.5 - 2|| Campylobacter rectus (ATCC 33238)| 1|| Campylobacter showae (ATCC 51146)| 0.5|| Capnocytophaga gingivalis (ATCC 33624)| 0.5|| Capnocytophaga ochracea (ATCC 27872)| 1|| Capnocytophaga spp.| 1|| Capnocytophaga sputigena (ATCC 33612)| 1|| Citrobacter diversus| >1000|| Citrobacter freundii| 2 - 2048|| Clostridium bifermentans| ≤0.125 - 0.5|| Clostridium cadaveris| ≤0.125 - 0.5|| Clostridium difficile| 1 - 4|| Clostridium histolyticum | ≤0.125 - 0.5|| Clostridium perfringens| ≤0.03 - 0.5|| Clostridium ramosum| ≤0.125 - 0.5|| Clostridium sordellii| ≤0.125 - 0.5|| Clostridium spiroforme| 0.032 - 0.5|| Clostridium spp.| ≤0.125 - 0.5|| Clostridium tertium| ≤0.125 - 0.5|| Corynebacterium matruchotii (ATCC 14266)| 0.5|| Corynebacterium spp. (HEGP 1006)| >256|| Edwardsiella hoshinae | 0.13 - 0.25|| Edwardsiella ictaluri | 0.13 - 1|| Edwardsiella tarda| 0.13 - 64|| Eikenella corrodens| 2 - 8|| Enterobacter cloacae| 4 - >500|| Enterobacteriaceae| 0.25 - 128|| Enterococci| 0.12 - 128|| Enterococcus faecalis| 0.25 - 7|| Enterococcus faecium| 32 - >256|| Escherichia coli| 2 - >500|| Eubacterium saburreum (ATCC 33271)| 0.5|| Eubacterium spp.| 1|| Eubacterium sulci (ATCC 35585)| 0.5|| Fusobacterium mortiferum| 1 - >128|| Fusobacterium necrophorum| ≤0.125 - 0.5|| Fusobacterium nucleatum| ≤0.125 - 16|| Fusobacterium periodonticum (ATCC 33693)| 32|| Fusobacterium spp.| ≤0.125 - >128|| Fusobacterium varium| 1 - 2|| Gemella morbillorum (ATCC 27824)| 0.5 - ?|| Haemolytic streptococci| 0.008 - 0.12|| Haemophilus ducreyi| 9|| Haemophilus influenzae| <0.07 - >64|| Haemophilus parasuis| 0.6 - 310|| Haemophilus spp.| 0.06 - 128|| Helicobacter bilis (ATCC 51630)| 4|| Helicobacter mustelae (ATCC 43772)| 4|| Helicobacter pullorum (ATCC 51864)| 4|| Helicobacter pylori| 0.0002 -8|| Klebsiella pneumonia| 2 - >500|| Lactobacillus acidophilus| ≤0.125 - 2|| Lactobacillus buchneri | 0.25 - 1|| Lactobacillus plantarum| ≤0.125 - 2|| Lactobacillus reuteri| ≤0.125 - 4|| Lactobacillus rhamnosus| 0.5 - 2|| Lactobacillus salivarius | ≤0.125 - 0.5|| Lactobacillus spp.| 0.5 - 129|| Lactococcus lactis| ≤0.125 - 1|| Leptotrichia buccalis (ATCC 14201)| 0.5|| Leuconostoc | ≤0.125 - 1|| Micrococcus| <0.24 - >40|| Moraxella catarrhalis (BC1)| <0.07|| Morganella morganii| >100 - 512|| Mycoplasma hyopneumoniae| 0.09 - 737.3|| Mycoplasma hyorhinis| 0.09 - 737.3|| Neisseria gonorrhoeae| 0.3|| Neisseria meningitidis (N2)| <0.07|| Neisseria mucosa (ATCC 19696)| 2|| Neisseria spp.| 0.004 - 32|| Nocardia asteroides| 6.2 - ≥400|| Ochrobactrum anthropi (SLO74)| ≥28|| Pasteurella multocida| 128 - 1240|| Pediococcus | ≤0.125 - 1|| Peptostreptococcus anaerobius| ≤0.125 - 32|| Peptostreptococcus asaccharolyticus| ≤0.125 - 32|| Peptostreptococcus magnus| ≤0.125 - 32|| Peptostreptococcus micros| ≤0.125 - 32|| Peptostreptococcus prevotii| ≤0.125 - 32|| Peptostreptococcus spp.| <1 - 8|| Peptostreptococcus tetradius| ≤0.125 - 32|| Pneumococci| 0.008 - 4|| Porphyromonas asaccharolytica| ≤0.0125 - 64|| Porphyromonas endodontalis (ATCC 35406)| 4|| Porphyromonas gingivalis| ≤0.0125 - 64|| Porphyromonas spp.| ≤0.125 - 128|| Prevotella bivia| ≤0.125 - 128|| Prevotella buccae| ≤0.0125 - 64|| Prevotella corporis | ≤0.0125 - 64|| Prevotella disiens| ≤0.0125 - 64|| Prevotella intermedia| ≤0.0125 - 64|| Prevotella loescheii| ≤0.0125 - 64|| Prevotella melaninogenica| ≤0.125 - 128|| Prevotella nigrescens| 0.5|| Prevotella oralis (group)| ≤0.0125 - 64|| Prevotella oris| ≤0.0125 - 64|| Prevotella spp.| ≤0.125 - 128|| Propionibacterium acnes| ≤0.125 - 0.25|| Proteus mirabilis (1287)| 0.39|| Proteus vulgaris| 1 - 1024|| Providencia rettgeri (NIH 96)| 3.13|| Pseudomonas aeruginosa| 1 - 1024|| Ralstonia pickettii| 64|| Salmonella spp.| 1|| Salmonella typhi| 6.25 - 32|| Selenomonas noxia| 0.5|| Serratia marcescens| 16 - >256 - ?|| Shigella flexneri| 2|| Staphylococci| 0.03 - 128|| Staphylococcus aureus| <0.0625 - >128|| Staphylococcus aureus (methicillin-resistant)| 0.2 - 512|| Staphylococcus epidermidis| 0.78 - 1|| Staphylococcus faecalis| 1.22 - 1.95|| Streptococcus agalactiae | 0.094|| Streptococcus anginosus| 0.5|| Streptococcus bovis| ≤0.125 - 1|| Streptococcus constellatus (ATCC 27823)| 0.5|| Streptococcus equi | 0.03 - 0.06|| Streptococcus faecalis (ATCC 29212)| 412|| Streptococcus gordonii (ATCC 10558)| 0.5|| Streptococcus infantarius | ≤0.125 - 1|| Streptococcus intermedius (ATCC 27335)| 1|| Streptococcus mitis (ATCC 49456)| 0.5|| Streptococcus oralis (ATCC 35037)| 0.5|| Streptococcus pneumonia| ≤0.008 - 31.25|| Streptococcus pyogenes| 32|| Streptococcus sanguinis| 1|| Streptococcus spp.| 32 - 128|| Streptococcus suis| 78 - 1240|| Tannerella forsythia| 8|| Veillonella parvula| 0.5|| Weissella spp.| ≤0.125 - 0.25|| Xanthomonas oryzae| 18|| |