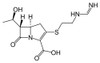
Imipenem is a broad-spectrum β-lactam antibiotic, a carbapenem effective against extended spectrum β-lactamase (ESBL) producing Enterobacteriaceae, a group of pathogenic microbes resistant to many first line β-lactam antibiotics and certain cephalosporins. Imipenem is a broad-spectrum antibiotic targeting a wide range of aerobic and anaerobic Gram-positive and Gram-negative bacteria. Imipenem is commonly used in susceptibility testing. Imipenem is sparingly soluble in water.
We also offer:
- Imipenem mixture w/cilastatin (I005)
| Mechanism of Action | β-lactams interfere with PBP (penicillin binding protein) activity involved in the final phase of peptidoglycan synthesis. PBP’s are enzymes which catalyze a pentaglycine crosslink between alanine and lysine residues providing additional strength to the cell wall. Without a pentaglycine crosslink, the integrity of the cell wall is severely compromised and ultimately leads to cell lysis and death. Resistance to β-lactams is commonly due to cells containing plasmid encoded β-lactamases. Like many members of the carbapenem subgroup, imipenem is highly resistant to β-lactamase activity. |
| Spectrum | Imipenem is a broad-spectrum antibiotic targeting a wide range of aerobic and anaerobic Gram-positive and Gram-negative bacteria. Resistance to Imipenem may emerge in Pseudomonas aeruginosa. |
| Microbiology Applications | Imipenem is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram-positive and Gram-negative microbial isolates. Imipenem has also shown high potency against high-resistant superbug strains. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
Microfluidic droplet systems are popular analytical methods used in antimicrobial susceptibility testing (AST) to help study antimicrobial resistance (AMR). Droplet methods have many advantages over conventional AST methods. Authors used several antibiotics from TOKU-E including Imipenem to review the physicochemical properties to predict retention of antibiotics in droplets (Ruszczak et al, 2023). |
| Plant Biology Applications | Imipenem has been shown to be effective against bacteria from the genus Burkholderia, a well described plant pathogen, and was shown to be effective for sour skin (onions), slippery skin (bulbs), and cavity disease (mushrooms)(Sojanova et al, 2007). |
| Molecular Formula | C12H17N3O4S · H2O |
| References |
Benjama A and Charkaoui B (1997) Control of Bacillus contaminating date palm tissue in micropropagation using antibiotics. In: Cassells AC (eds) Pathogen and microbial contamination management in micropropagation. Developments in plant pathology. Vol 12 Springer, Dordrecht. pp 207-211 Berg PH, Voit EO and White RL (1996) A pharmacodynamic model for the action of the antibiotic Imipenem on Pseudomonas aeruginosa populations in vitro. Bltn. Mathcal. Biol. 58(5):923-938 Hellinger WC, Brewer NS (1991) Imipenem. Mayo Clinic Proc. 66(10):1074-1081 Pitout JD, Sanders CC, Sanders WE (1997) Antimicrobial resistance with focus on beta-lactam resistance in Gram-negative bacilli. Am J Med 103:51 Ruszczak A, Jankowski P, Vasantham S, Scheler O and Garstiecki P (2023) Physicochemical properties predict retention of antibiotics in water-in-oil droplets. Anal. Chem. 95(2): 1574−1581 PMID 36598882 |
| MIC | Achromobacter xylosoxidans subsp. denitrificans| 0.25 - 4 || Acinetobacter anitratus| ≤0.008 - 128 || Acinetobacter baumannii| 0.008 - 512 || Acinetobacter calcoaceticus| 0.016 - >8 || Acinetobacter haemolyticus| ≤0.008 - >16 || Acinetobacter junii| ≤0.12 - >8 || Acinetobacter lwoffii| ≤0.008 - >16 || Acinetobacter spp.| 0.008 - >64 || Actinomyces gerencseriae| ≤0.015 - 8 || Actinomyces graevenitzii | ≤0.015 - 0.25 || Actinomyces israelii| ≤0.015 - 8 || Actinomyces meyeri| ≤0.015 - 8 || Actinomyces naeslundii| 0.015 - 8 || Actinomyces neuii| ≤0.015 - 0.25 || Actinomyces odontolyticus| ≤0.015 - 8 || Actinomyces radingae| ≤0.015 - 0.25 || Actinomyces schalii| ≤0.015 - 0.25 || Actinomyces spp.| ≤0.008 - 8 || Actinomyces turicensis| ≤0.015 - 0.25 || Actinomyces viscosus| ≤0.015 - 0.5 || Aerococcus spp.| ≤0.008 - 4 || Aerococcus urinae| ≤0.008 - 4 || Aeromonas caviae| 0.25 - 4 || Aeromonas hydrophila| 0.25 - 16 || Aeromonas spp.| 0.12 - 4 || Agrobacterium radiobacter| 0.06 - 1 || Alcaligenes faecalis| 0.06 - >16 || Alcaligenes odorans| 0.25 - 1 || Anaerococcus prevotii| ≤0.016 - 0.25 || Anaerococcus tetradius | ≤0.016 - 0.03 || Arcanobacterium pyogenes| ≤0.03 - 0.25 || Atopobium parvulum| 0.25|| Bacillus proteus| 4|| Bacillus spp.| ≤0.008 - 4 || Bacillus subtilis| <0.025 || Bacteroides caccae| ≤0.06 - 8 || Bacteroides capillosus | 0.06 - 0.25 || Bacteroides distasonis| 0.03 - 8 || Bacteroides eggerthii| ≤0.125 - 0.5 || Bacteroides fragilis| ≤0.008 - >128 || Bacteroides fragilis gr.| 0.03 - 4 || Bacteroides levii| 0.06 - 0.25 || Bacteroides merdae| ≤0.06 - 4 || Bacteroides ovatus| 0.03 - 16 || Bacteroides splanchnicus | 0.06 - 0.25 || Bacteroides spp.| ≤0.004 - 4 || Bacteroides stercoris| 0.06 - 8 || Bacteroides thetaiotaomicron| 0.03 - >128 || Bacteroides uniformis| 0.03 - 4 || Bacteroides ureolyticus| 0.06 - 0.25 || Bacteroides vulgatus| 0.03 - 16 || Bifidobacterium adolescentis| ≤0.03 - 1 || Bifidobacterium bifidum| ≤0.03 - 0.25 || Bifidobacterium breve| ≤0.03 - 2 || Bifidobacterium catenulatum| ≤0.03 - 0.25 || Bifidobacterium dentium| ≤0.03 - 0.25 || Bifidobacterium longum| ≤0.03 - 8 || Bifidobacterium pseudolongum| 8|| Bifidobacterium spp.| ≤0.008 - 2 || Bilophila wadsworthia| 0.06 - 0.5 || Bordetella bronchiseptica| 0.06 - >16 || Bordetella pertussis | 0.25 - 4 || Branhamella catarrhalis| 0.015 - 4 || Brevibacterium spp.| ≤0.03 - >32 || Brevundimonas vesicularis| 0.06 - >64 || Burkholderia cepacia| 0.06 - 256 || Burkholderia spp.| 16 - >32 || Campylobacter fetus| ≤0.06 - 0.25 || Campylobacter gracilis| 0.06 - 0.5 || Campylobacter jejuni| ≤0.008 - 0.12 || Campylobacter pylori| ≤0.008 - 0.015 || Candida albicans| >25000|| Capnocytophaga ochracea | 0.5|| Chryseobacterium indologenes| ≤0.06 - >64 || Chryseobacterium meningosepticum| 32 - 64 || Citrobacter amalonaticus| 0.006 - >8 || Citrobacter braakii| 0.06 - >8 || Citrobacter diversus| 0.06 - >8 || Citrobacter farmeri| 0.06 - >8 || Citrobacter freundii| 0.03 - 16 || Citrobacter koseri| 0.06 - >8 || Citrobacter spp.| 0.06 - >8 || Clostridium aminovalericum| ≤0.03 - 1 || Clostridium baratii| ≤0.03 - 4 || Clostridium bifermentans| ≤0.015 - 4 || Clostridium butyricum| ≤0.015 - 2 || Clostridium cadaveris| ≤0.015 - 4 || Clostridium clostridioforme| ≤0.03 - 4 || Clostridium cochlearium| ≤0.015 - 2 || Clostridium difficile| ≤0.06 - 64 || Clostridium fallax| ≤0.015 - 2 || Clostridium glycolicum| ≤0.015 - 2 || Clostridium hastiforme| 0.06 - 4 || Clostridium histolyticum | 0.06 - 0.5 || Clostridium innocuum| 0.06 - 4 || Clostridium leptum| ≤0.03 - 1 || Clostridium novyi| ≤0.015 - 2 || Clostridium oroticum| ≤0.015 - 2 || Clostridium paraputrificum| ≤0.015 - 2 || Clostridium perfringens| 0.008 - 1 || Clostridium putrificum| 0.125|| Clostridium ramosum| 0.03 - 4 || Clostridium septicum| ≤0.03 - 1 || Clostridium sordellii| ≤0.03 - 4 || Clostridium sphenoides| ≤0.03 - 1 || Clostridium sporogenes| ≤0.03 - 1 || Clostridium spp.| ≤0.004 - 8 || Clostridium subterminale| ≤0.03 - 1 || Clostridium symbiosum| ≤0.03 - 1 || Clostridium tertium| 0.06 - 4 || Collinsella aerofaciens| ≤0.03 - 0.25 || Comamonas acidovorans| 0.25 - 4 || Comamonas spp.| 0.06 - >16 || Corynebacterium| ≤0.015 - >32 || Corynebacterium accolens| ≤0.015 - 4 || Corynebacterium afermentans| ≤0.008 - >32 || Corynebacterium amycolatum| ≤0.008 - >32 || Corynebacterium aquaticum| ≤0.015 - >32 || Corynebacterium falsenii| ≤0.015 - 4 || Corynebacterium jeikeium| ≤0.008 - >32 || Corynebacterium minutissimum| ≤0.015 - >32 || Corynebacterium pseudodiphtheriticum| ≤0.008 - >32 || Corynebacterium spp.| ≤0.007 - >8 || Corynebacterium striatum| ≤0.008 - >16 || Corynebacterium urealyticum| ≤0.008 - >32 || Corynebacterium xerosis| ≤0.03 - >32 || Dermabacter hominis| ≤0.015 - 4 || Desulfovibrio piger | 0.25|| Eggerthella lenta| 0.5 - 4 || Eggerthella lenta| 8|| Eikenella corrodens| ≤0.12 - 1 || Enterobacter aerogenes| 0.125 - 96 || Enterobacter agglomerans| 0.125 - >16 || Enterobacter amnigenus| ≤0.5 - >8 || Enterobacter asburiae| 0.12 - >16 || Enterobacter cancerogenus| 0.12 - >16 || Enterobacter cancerogenus| 0.5|| Enterobacter cancerogenus| 0.12|| Enterobacter cloacae| 0.06 - 97 || Enterobacter gergoviae| ≤0.5 - >8 || Enterobacter hormaechei| ≤0.5 - >8 || Enterobacter intermedius | 0.12 - >16 || Enterobacter sakazakii| 0.125 - >16 || Enterobacter spp.| ≤0.12 - >16 || Enterobacter taylorae| ≤0.5 - >8 || Enterobacteriaceae| ≤0.06 - >16 || Enterococci| 0.25 - 128 || Enterococcus avium| ≤0.03 - >64 || Enterococcus casseliflavus| 0.03 - >16 || Enterococcus durans| 0.03 - >16 || Enterococcus faecalis| 0.03 - ≥128 || Enterococcus faecium| ≤0.03 - >128 || Enterococcus gallinarum| 0.03 - >16 || Enterococcus hirae| ≤1 - >8 || Enterococcus raffinosus| 0.03 - >16 || Enterococcus spp.| 0.03 - >16 || Escherichia coli| 0.006 - >99 || Eubacterium alactolyticum| ≤0.03 - 0.25 || Eubacterium brachy| ≤0.015 - 0.5 || Eubacterium combesii| ≤0.015 - 0.5 || Eubacterium contortum| ≤0.015 - 0.5 || Eubacterium lentum| ≤0.015 - 4 || Eubacterium limosum| ≤0.015 - 0.5 || Eubacterium moniliforme | ≤0.03 - 0.125 || Eubacterium saburreum| ≤0.015 - 0.5 || Eubacterium spp.| ≤0.016 - 0.5 || Eubacterium tenue| ≤0.015 - 0.5 || Eubacterium timidum| ≤0.015 - 0.5 || Eubacterium yurii| ≤0.015 - 0.5 || Finegoldia magna| ≤0.03 - 0.125 || Flavobacterium breve| 0.06 - >16 || Flavobacterium mengosepticum| 0.06 - >16 || Fusobacteria| ≤0.016 - 1 || Fusobacterium mortiferum| 0.06 - 1.5 || Fusobacterium necrophorum| ≤0.0016 - 4 || Fusobacterium nucleatum| ≤0.016 - 4 || Fusobacterium spp.| ≤0.008 - 4 || Fusobacterium varium| 0.06 - 4 || Gemella morbillorum| ≤0.03 - 0.5 || Gram-Positive Anaerobic Cocci| 0.03 - 0.12 || Haemolytic streptococci| 0.002 - 0.25 || Haemophilus aegyptius| ≤0.008 - 2 || Haemophilus aphrophilus| ≤0.008 - 2 || Haemophilus haemolyticus| ≤0.008 - 2 || Haemophilus influenzae| ≤0.008 - 64 || Haemophilus parahaemolyticus| ≤0.008 - 2 || Haemophilus parainfluenzae| ≤0.008 - 2 || Haemophilus spp.| 0.25 - 4 || Hafnia alvei| 0.06 - >16 || Helicobacter pylori| 0.05|| Klebsiella aerogenes| 0.2|| Klebsiella baumannii| 4|| Klebsiella ornithinolytica| 0.06 - >16 || Klebsiella ozaenae| ≤0.5 - >8 || Klebsiella pneumonia| 0.03 - 6250|| Klebsiella spp.| 0.06 - >8 || Klebsiella terrigena| ≤0.5 - >8 || Lactobacillus acidophilus| ≤0.015 - 8 || Lactobacillus brevis| 0.015 - ≤8 || Lactobacillus casei| ≤0.015 - 16 || Lactobacillus catenaforme| ≤0.015 - 8 || Lactobacillus confusus| ≤0.03 - 8 || Lactobacillus delbrueckii| ≤0.03 - 8 || Lactobacillus fermentans| ≤0.015 - 8 || Lactobacillus fermentum| 0.03|| Lactobacillus fermentum| 0.03|| Lactobacillus gasseri| ≤0.03 - >100 || Lactobacillus jensenii| ≤0.015- 8 || Lactobacillus johnsonii| >100 || Lactobacillus lactis| ≤0.03 - 8 || Lactobacillus leichmannii | ≤0.03 - 1 || Lactobacillus minutus| ≤0.015 - 8 || Lactobacillus oris| ≤0.015 - 8 || Lactobacillus paracasei| >100 || Lactobacillus plantarum| ≤0.015 - 8 || Lactobacillus reuteri| 0.06|| Lactobacillus rhamnosus| ≤0.015 - 8 || Lactobacillus salivarius | 1 - 10|| Lactobacillus spp.| ≤0.008 - 4|| Lactobacillus uli | ≤0.03 - 1|| Legionella pneumophila| ≥0.063|| Leuconostoc spp.| >1000|| Listeria monocytogenes| ≤0.008 - 4|| Micrococcus spp.| ≤0.008 - 4|| Micromonas micros| 0.015 - 2|| Moraxella catarrhalis| ≤0.008 - 1|| Morganella morganii| 0.016 - 16|| Morganella spp.| ≤0.12 - 16|| Mycobacterium avium-intracellulare complex| 2 - 32|| Neisseria meningitidis| 0.03 - 1|| Neisseria meningitidis| 0.03|| Neisseria spp.| 0.004 - 0.25|| Nonsporing gram-positive rods| 0.03 - 1|| Ochrobactrum anthropi| 1 - 2|| Oligella ureolytica| ≤0.06 - >64|| Pandoraea apista| <1 - 4|| Pandoraea pnomenusa| <1 - 2|| Pantoea agglomerans| ≤1 - 2|| Pasteurella multocida| 0.03 - 1|| Pasteurella spp.| 0.06 - >16|| Pemphigus mirabilis| 4|| Pemphigus vulgaris| 4|| Peptococcus spp.| 0.004 - 0.5|| Peptoniphilus asaccharolyticus| ≤0.016 - 1|| Peptoniphilus indolicus| 0.5|| Peptostreptococcus anaerobius| 0.004 - 32|| Peptostreptococcus asaccharolyticus| ≤0.015 - 2|| Peptostreptococcus magnus| ≤0.03 - 32|| Peptostreptococcus micros| ≤0.016 - 2|| Peptostreptococcus prevotii| ≤0.015 - 2|| Peptostreptococcus spp.| ≤0.004 - 32|| Peptostreptococcus tetradius| ≤0.016 - 2|| Plesiomonas shigelloides| 0.25 - 1|| Pneumococci| 0.002 - 0.5|| Porphyromonas asaccharolytica| ≤0.016 - 0.25|| Porphyromonas endodontalis | 0.06 - 0.25|| Porphyromonas gingivalis| ≤0.016 - 0.25|| Porphyromonas levii | ≤0.016|| Porphyromonas spp.| ≤0.016 - 1|| Prevotella bivia| ≤0.016 - 2|| Prevotella buccae| ≤0.016 - 4|| Prevotella corporis | ≤0.016 - 0.125|| Prevotella denticola| ≤0.016 - 0.125|| Prevotella disiens| ≤0.016 - 2|| Prevotella heparinolytica | 0.125|| Prevotella intermedia| ≤0.016 - 4|| Prevotella loescheii| ≤0.016 - 0.25|| Prevotella melaninogenica| ≤0.016 - 0.25|| Prevotella melaninogenicus | 0.06 - 4|| Prevotella nigrescens| ≤0.016 - 0.25|| Prevotella oralis| ≤0.016 - 4 || Prevotella oris| ≤0.016 - 0.5|| Prevotella spp.| ≤0.008 - 4|| Prevotella/Porphyromonas spp.| ≤0.016 - 0.125|| Propionibacterium spp.| ≤0.008 - 0.5|| Propionibacterium acnes | ≤0.008 - 1|| Propionibacterium avidum| ≤0.015 - 0.06|| Propionibacterium granulosum| ≤0.015 - 0.125|| Propionibacterium propionicus| ≤0.015 - 0.06|| Proteus mirabilis| 0.016|| Proteus rettgeri| 0.016 - 16|| Proteus spp.| 0.5 - 8|| Proteus vulgaris| 0.016 - 16|| Providencia alcalifaciens| 41290|| Providencia rettgeri| 0.39 - 32|| Providencia spp.| ≤0.12 - >32|| Providencia stuartii| 0.016 - 16|| Pseudomonas aeruginosa| 0.12 - 64000 μg/ml|| Pseudomonas cepacia| 0.25 - >128|| Pseudomonas fluorescens | 0.12 - >16|| Pseudomonas oryzihabitans| 0.13 - 2|| Pseudomonas paucimobilis| 0.12 - >16|| Pseudomonas putida| 0.12 - >16|| Pseudomonas spp.| 0.06 - >16|| Pseudomonas stutzeri| 0.06 - >16|| Pseudomonas vesicularis| 0.12 - >16|| Pseudoramibacter alactolyticus | ≤0.03 - 0.125|| Ralstonia pickettii | ≤0.06 - 2|| Rothia spp.| <0.015 - 4|| Salmonella agona| ≤0.5 - 2|| Salmonella arizonae| ≤0.5 - 2|| Salmonella bareilly| ≤0.5 - 2|| Salmonella enterica| 0.12 - 2|| Salmonella enteritidis| 0.125 - 2|| Salmonella hadar| ≤0.5 - 2|| Salmonella heidelberg| ≤0.5 - 2|| Salmonella infantis| ≤0.5 - 2|| Salmonella litchfield| ≤0.5 - 2|| Salmonella montevideo| ≤0.5 - 2|| Salmonella muenchen| ≤0.5 - 2|| Salmonella newport| ≤0.5 - 2|| Salmonella panama| ≤0.5 - 2|| Salmonella Paratyphi| ≤0.5 - 2|| Salmonella schwarzengrund| ≤0.5 - 2|| Salmonella spp.| 0.12 - 2|| Salmonella stpaul| ≤0.5 - 2|| Salmonella thompson| ≤0.5 - 2|| Salmonella typhi| 0.12 - 2|| Salmonella virchow| ≤0.5 - 2|| Serratia fonticola| ≤0.12 - >8|| Serratia liquefaciens| 0.06 - >16|| Serratia marcescens| 0.12 - >100|| Serratia odorifera| ≤0.12 - >8|| Serratia plymuthica| ≤0.12 - >8|| Serratia rubidaea| 0.06 - >16|| Serratia spp.| ≤0.12 - 16|| Shewanella putrefaciens| 0.06 - >16|| Shigella boydii| 0.06 - ≤0.5|| Shigella dysenteriae| ≤0.5|| Shigella flexneri| 0.06 - 2|| Shigella sonnei| 0.06 - 2|| Shigella spp.| 0.06 - ≤0.5|| Shingella dysenteriae| 0.125 - 2|| Sphingomonas spp.| 0.5 - >64|| Staphylococcus aureus| 0.004 - 195.31|| Staphylococcus auricularis| ≤0.5 - >8|| Staphylococcus capitis| ≤0.5 - >8|| Staphylococcus caprae| ≤0.5 - >8|| Staphylococcus cohnii| ≤0.5 - >8|| Staphylococcus epidermidis| ≤0.004 - 128|| Staphylococcus haemolyticus| ≤0.007 - >128|| Staphylococcus hominis| ≤0.007 - >128|| Staphylococcus intermedius| ≤0.5 - >8|| Staphylococcus lugdunensis| ≤0.5 - >8|| Staphylococcus mitis| ≤0.007 - 1|| Staphylococcus saccharolyticus| ≤0.03|| Staphylococcus sanguis| 0.007 - ≤0.25|| Staphylococcus saprophyticus| 0.015 - >8|| Staphylococcus sciuri| ≤0.5 - >8|| Staphylococcus simulans| ≤0.007 - >8|| Staphylococcus spp.| ≤0.007 - 256|| Staphylococcus warneri| ≤0.5 - >8|| Stenotrophomonas maltophilia| 4 - 512|| Streptococcus agalactiae| 0.008 - 64|| Streptococcus bovis| ≤0.008 - 2|| Streptococcus constellatus| 0.125|| Streptococcus intermedius| 0.5|| Streptococcus milleri| ≤0.008 - 2|| Streptococcus mutans| ≤0.008 - 2|| Streptococcus pneumoniae| 0.002 - 2|| Streptococcus pyogenes| 0.002 - 0.125|| Streptococcus spp.| 0.002 - 288|| Sutterella wadsworthensis| 0.03 - 16|| Veillonella dispar | 0.06|| Veillonella parvula | ≤0.008 - 4|| Veillonella spp.| 0.015 - 4|| Weeksella virosa | ≤0.06 - 0.13|| Xanthomonas maltophilia| 64 - 128|| Yersinia enterocolitica| 0.06 - 16|| |